mattest
Two-sample t-test to evaluate differential expression of genes from two experimental conditions or phenotypes
Syntax
Description
pvalues = mattest(xdata,ydata)xdata and
ydata. The function returns a
p-value for each gene.
xdata contains data from one experimental
condition and ydata contains data from
another experimental condition. For example,
xdata could be expression
values from cancer cells, and ydata
could be expression values from normal cells.
xdata and ydata must
have the same number of rows and are assumed to be normally
distributed in each class with equal variances.
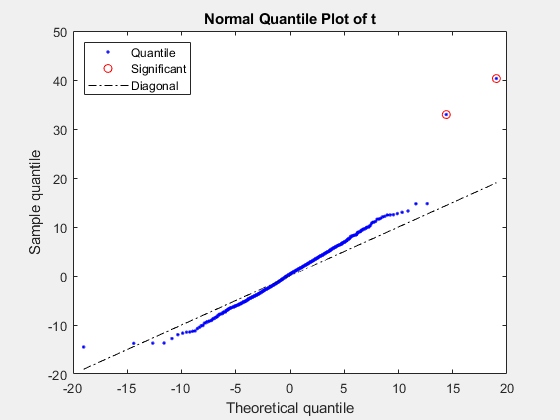
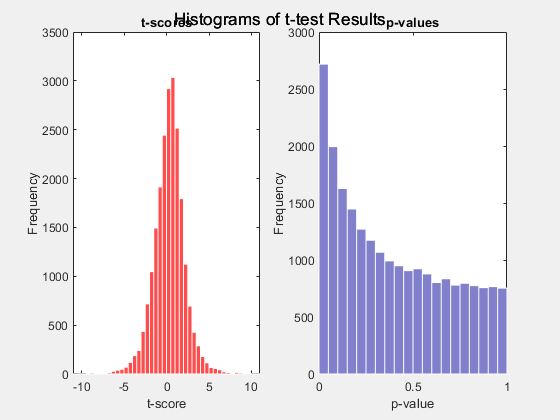
pvalues = mattest(xdata,ydata,Name=Value)ShowPlot to
true.
Examples
Input Arguments
Name-Value Arguments
Output Arguments
References
[1] Review Literature: Huber, W., von Heydebreck, A., Sültmann, H., Poustka, A., and Vingron, M. (2002). Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics 18 (Suppl. 1), S96–S104.
[2] Best, C.J.M., Gillespie, J.W., Yi, Y., Chandramouli, G.V.R., Perlmutter, M.A., Gathright, Y., Erickson, H.S., Georgevich, L., Tangrea, M.A., Duray, P.H., Gonzalez, S., Velasco, A., Linehan, W.M., Matusik, R.J., Price, D.K., Figg, W.D., Emmert-Buck, M.R., and Chuaqui, R.F. (2005). Molecular alterations in primary prostate cancer after androgen ablation therapy. Clinical Cancer Research 11, 6823–6834.
Version History
Introduced in R2006a
See Also
maboxplot | mafdr | mainvarsetnorm | mairplot | maloglog | malowess | manorm | mavolcanoplot