detect
Description
[
detects anomalies in signals stored in lbls,loss] = detect(d,data)data.
The function assigns a normal label to signal windows whose aggregated loss value is below the detection threshold, and an abnormal label to signal windows whose aggregated loss value is greater than or equal to the detection threshold.
Examples
Load a convolutional anomaly detector trained with three-channel sinusoidal signals. Display the model, threshold, and window properties of the detector.
load sineWaveAnomalyDetector
DD =
deepSignalAnomalyDetectorCNN with properties:
IsTrained: 1
NumChannels: 3
Model Information
ModelType: 'convautoencoder'
FilterSize: 8
NumFilters: 32
NumDownsampleLayers: 2
DownsampleFactor: 2
DropoutProbability: 0.2000
Threshold Information
Threshold: 0.0510
ThresholdMethod: 'contaminationFraction'
ThresholdParameter: 0.0100
Window Information
WindowLength: 1
OverlapLength: 'auto'
WindowLossAggregation: 'mean'
Load the file sineWaveAnomalyData.mat, which contains two sets of synthetic three-channel sinusoidal signals.
sineWaveNormalcontains the 10 sinusoids used to train the convolutional anomaly detector. Each signal has a series of small-amplitude impact-like imperfections but otherwise has stable amplitude and frequency.sineWaveAbnormalcontains three signals of similar length and amplitude to the training data. One of the signals has an abrupt, finite-time change in frequency. Another signal has a finite-duration amplitude change in one of its channels. A third has random spikes in each channel.
Plot three normal signals and the three signals with anomalies.
load sineWaveAnomalyData tiledlayout(3,2,TileSpacing="compact",Padding="compact") rnd = randperm(length(sineWaveNormal)); for kj = 1:3 nexttile plot(sineWaveNormal{rnd(kj)}) title("Normal Signal") nexttile plot(sineWaveAbnormal{kj}) title("Signal with Anomalies") end
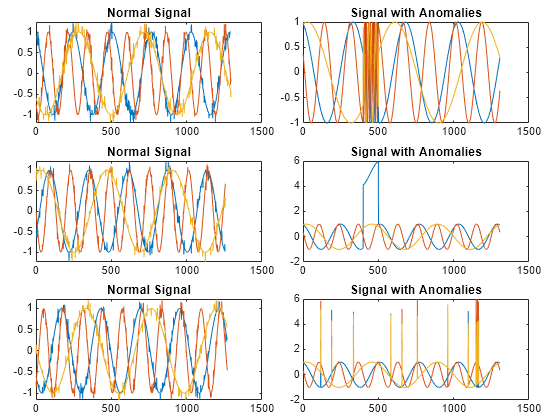
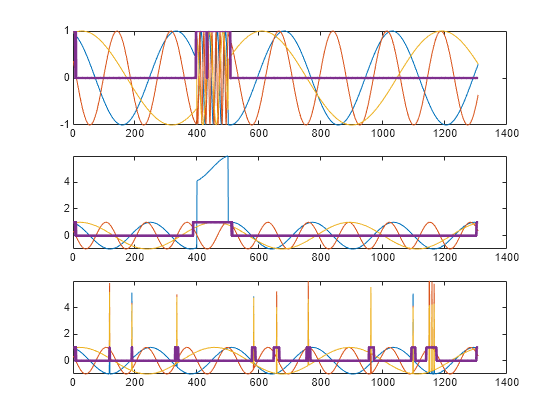
Use the trained anomaly detector to detect the anomalies in the abnormal data.
[lbls,loss] = detect(D,sineWaveAbnormal);
The first output of detect is a categorical array that declares each sample of a signal as being anomalous or not.
tiledlayout("vertical") for kj = 1:3 nexttile plot(sineWaveAbnormal{kj}) hold on plot(lbls{kj},LineWidth=2) end
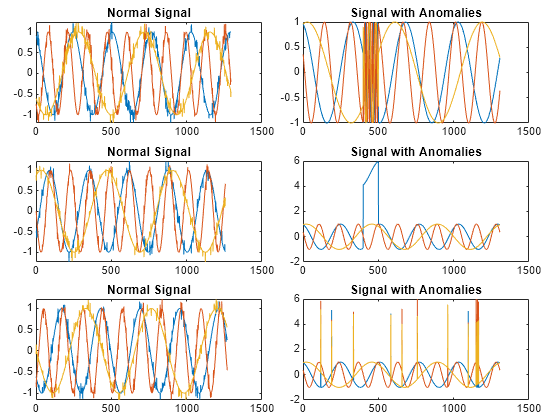
Load the file sineWaveAnomalyData.mat, which contains two sets of synthetic three-channel sinusoidal signals.
sineWaveNormalcontains the 10 sinusoids used to train the convolutional anomaly detector. Each signal has a series of small-amplitude impact-like imperfections but otherwise has stable amplitude and frequency.sineWaveAbnormalcontains three signals of similar length and amplitude to the training data. One of the signals has an abrupt, finite-time change in frequency. Another signal has a finite-duration amplitude change in one of its channels. A third has random spikes in each channel.
Plot three normal signals and the three signals with anomalies.
load sineWaveAnomalyData tiledlayout(3,2,TileSpacing="compact",Padding="compact") rnd = randperm(length(sineWaveNormal)); for kj = 1:length(sineWaveAbnormal) nexttile plot(sineWaveNormal{rnd(kj)}) title("Normal Signal") nexttile plot(sineWaveAbnormal{kj}) title("Signal with Anomalies") end
Create a long short-term memory (LSTM) forecaster object to detect the anomalies in the abnormal signals. Specify a window length of 10 samples.
D = deepSignalAnomalyDetector(3,"lstmforecaster",windowLength=10);Train the forecaster using the anomaly-free sinusoids. Use the training options for the adaptive moment estimation (Adam) optimizer and specify a maximum number of 100 epochs. For more information, see trainingOptions (Deep Learning Toolbox).
opts = trainingOptions("adam",MaxEpochs=100,ExecutionEnvironment="cpu"); trainDetector(D,sineWaveNormal,opts)
Iteration Epoch TimeElapsed LearnRate TrainingLoss
_________ _____ ___________ _________ ____________
1 1 00:00:00 0.001 0.6369
50 50 00:00:02 0.001 0.19706
100 100 00:00:03 0.001 0.064225
Training stopped: Max epochs completed
Computing threshold...
Threshold computation completed.
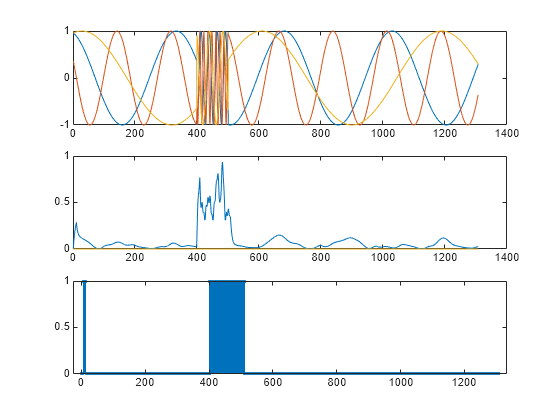
Use the trained detector to find the anomalies in the first signal. Reset the state of the detector. Stream the data one sample at a time and have the detector keep its state after each reading. Compute the reconstruction loss for each one-sample frame. Categorize signal regions where the loss exceeds a specified threshold as anomalous.
resetState(D)
sg = sineWaveAbnormal{1};
anoms = NaN(size(sg));
losss = zeros(size(sg));
for kj = 1:length(sg)
frame = sg(kj,:);
[lb,lo] = detect(D,frame, ...
KeepState=true,ExecutionEnvironment="cpu");
anoms(kj) = lb;
losss(kj) = lo;
endPlot the anomalous signal, the reconstruction loss, and the categorical array that declares each sample of the signal as being anomalous or not.
figure tiledlayout("vertical") nexttile plot(sg) nexttile plot(losss) nexttile stem(anoms,".")
Reset the state of the detector. Find the anomalies in the third signal. Plot the anomalous signal, the reconstruction loss, and the categorical array that declares each sample of the signal as being anomalous or not.
resetState(D)
sg = sineWaveAbnormal{3};
anoms = NaN(size(sg));
losss = zeros(size(sg));
for kj = 1:length(sg)
frame = sg(kj,:);
[lb,lo] = detect(D,frame, ...
KeepState=true,ExecutionEnvironment="cpu");
anoms(kj) = lb;
losss(kj) = lo;
end
figure
tiledlayout("vertical")
nexttile
plot(sg)
nexttile
plot(losss)
nexttile
stem(anoms,".")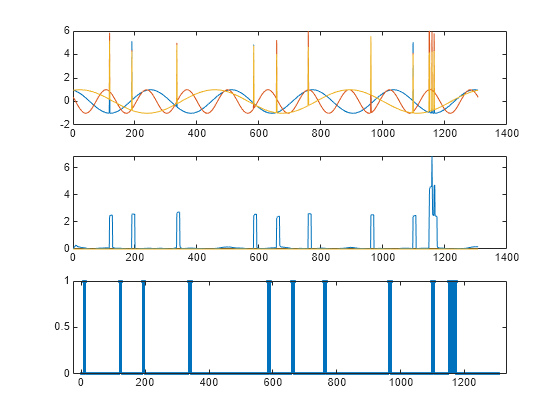
Input Arguments
Anomaly detector, specified as a deepSignalAnomalyDetectorCNN object, a
deepSignalAnomalyDetectorLSTM object, or a deepSignalAnomalyDetectorLSTMForecaster object. Use the
deepSignalAnomalyDetector function to create
d.
Signal data set, specified as one of these:
Nc-column matrix — A single multichannel signal observation (M = 1), where Nc is equal to the value of the
NumChannelsproperty of the detector.M-element cell array — M multichannel signal observations, where each cell contains an Nc-column matrix.
Timetable — A single multichannel signal observation, contained in a MATLAB® timetable. The timetable must contain increasing, uniformly-sampled, and finite values. The timetable can have:
A single variable containing an Nc-column matrix, where each column corresponds to a signal channel.
Nc variables, where each variable contains a vector that corresponds to a signal channel.
Datastore — A
signalDatastore,audioDatastore(Audio Toolbox), orarrayDatastoreobject. The detector uses thereadallfunction to read all the signal observations contained in the datastore at once. You can also use aCombinedDatastoreorTransformedDatastoreobject containing any of the supported datastores.
Name-Value Arguments
Specify optional pairs of arguments as
Name1=Value1,...,NameN=ValueN, where Name is
the argument name and Value is the corresponding value.
Name-value arguments must appear after other arguments, but the order of the
pairs does not matter.
Example: MiniBatchSize=64,ExecutionEnvironment="cpu" instructs the
function to use a mini-batch size of 64 and use the computer CPU to detect
anomalies.
Option to expand window labels to each sample, specified as either
false or true.
If you specify
ExpandWindowLabelsasfalse, thenlblscontains one label per window.If you specify
ExpandWindowLabelsastrue, thenlblscontains one label per sample.
Data Types: logical
Label priority for overlapped windows when ExpandWindowLabels
is true, specified as "anomaly" or
"normal".
If you specify
OverlapPriorityas"anomaly", the function labels samples in an abnormal window overlapped with a normal window as abnormal.If you specify
OverlapPriorityas"normal", the function labels samples in a normal window overlapped with an abnormal window as normal.
This argument applies only when you set
ExpandWindowLabels is true and the window
overlap length is greater than zero.
Data Types: char | string
Since R2024a
Option to keep the internal states of the anomaly detector so they persist during
subsequent calls to detect, specified as
false or true. Specify this argument as
true when processing a continuous signal across multiple calls to
ensure that the model incorporates historical data.
KeepState can be true only when:
ModelTypeis"lstmforecaster".WindowLengthis a number.fullSignalis not supported.OverlapLengthis equal toWindowLength– 1.datahas a batch size greater than 1.
Data Types: logical
Mini-batch size used by the network to compute reconstructed signals, specified as a positive integer scalar.
Data Types: single | double | int8 | int16 | int32 | int64 | uint8 | uint16 | uint32 | uint64
Execution environment used by the network, specified as one of these:
"auto"— If available, use the GPU. If the GPU is not available, use the CPU."gpu"— Use the GPU."cpu"— Use the CPU.
Data Types: char | string
Output Arguments
Labels, returned as a logical column vector, cell array, or timetable. The output
format of lbls depends on the format of data.
If
datais an Nc-column matrix, thenlblsis a logical column vector.If
datais an M-element cell array, thenlblsis a cell array containing M logical column vectors.If
datais a timetable, thenlblsis a timetable with a single variable containing a logical column vector.If
datais a datastore, thenlblsis a vector or cell array depending on the format of the data contained in the datastore.
A normal label has a value equal to false, and an abnormal
label has a value equal to true.
Window loss, returned as a column vector, cell array, or timetable. The output
format of loss depends on the format of data:
If
datais an Nc-column matrix, thenlossis a single-precision column vector.If
datais an M-element cell array, thenlossis a cell array containing M single-precision column vectors.If
datais a timetable, thenlossis a timetable with a single variable containing a single-precision column vector.If
datais a datastore, thenlossis a vector or cell array depending on the format of the data contained in the datastore.
Extended Capabilities
The detect function
supports GPU array input with these usage notes and limitations:
The ExecutionEnvironment option must be "gpu" or
"auto" when the input data is:
A
gpuArrayA cell array containing
gpuArrayobjectsA datastore that outputs cell arrays containing
gpuArrayobjects
For more information, see Run MATLAB Functions on a GPU (Parallel Computing Toolbox).
Version History
Introduced in R2023aYou have the option to keep the internal states of the anomaly detector so they persist
during subsequent calls to detect. Specify the
KeepState name-value argument as true when
processing a continuous signal across multiple calls to ensure that the model incorporates
historical data.
See Also
Objects
deepSignalAnomalyDetectorCNN|deepSignalAnomalyDetectorLSTM|deepSignalAnomalyDetectorLSTMForecaster
Functions
MATLAB Command
You clicked a link that corresponds to this MATLAB command:
Run the command by entering it in the MATLAB Command Window. Web browsers do not support MATLAB commands.
Select a Web Site
Choose a web site to get translated content where available and see local events and offers. Based on your location, we recommend that you select: .
You can also select a web site from the following list
How to Get Best Site Performance
Select the China site (in Chinese or English) for best site performance. Other MathWorks country sites are not optimized for visits from your location.
Americas
- América Latina (Español)
- Canada (English)
- United States (English)
Europe
- Belgium (English)
- Denmark (English)
- Deutschland (Deutsch)
- España (Español)
- Finland (English)
- France (Français)
- Ireland (English)
- Italia (Italiano)
- Luxembourg (English)
- Netherlands (English)
- Norway (English)
- Österreich (Deutsch)
- Portugal (English)
- Sweden (English)
- Switzerland
- United Kingdom (English)