gradient
(Not recommended) Evaluate gradient of function approximator object given observation and action input data
Since R2022a
gradient is not recommended. Use dlgradient and
dlfeval on your
loss function instead. For more information, see gradient is not recommended.
Syntax
Description
Examples
Create observation and action specification objects (or
alternatively use getObservationInfo and
getActionInfo to extract the specification objects from an
environment). For this example, define an observation space with three channels. The
first channel carries an observation from a continuous three-dimensional space, so that
a single observation is a column vector containing three doubles. The second channel
carries a discrete observation made of a two-dimensional row vector that can take one of
five different values. The third channel carries a discrete scalar observation that can
be either zero or one. Finally, the action space is a continuous four-dimensional space,
so that a single action is a column vector containing four doubles, each between
-10 and 10.
obsInfo = [
rlNumericSpec([3 1])
rlFiniteSetSpec({[1 2],[3 4],[5 6],[7 8],[9 10]})
rlFiniteSetSpec([0 1])
];
actInfo = rlNumericSpec([4 1], ...
UpperLimit= 10*ones(4,1), ...
LowerLimit=-10*ones(4,1) );To approximate the policy within the actor, use a recurrent deep neural network. For a continuous Gaussian actor, the network must have two output layers (one for the mean values the other for the standard deviation values), each having as many elements as the dimension of the action space.
Create a the network, defining each path as an array of layer objects. Use
sequenceInputLayer as the input layer and include an
lstmLayer as one of the other network layers. Also use a softplus
layer to enforce nonnegativity of the standard deviations and a ReLU layer to scale the
mean values to the desired output range. Get the dimensions of the observation and
action spaces from the environment specification objects, and specify a name for the
input layers, so you can later explicitly associate them with the appropriate
environment channel.
obs1Path = [
sequenceInputLayer( ...
prod(obsInfo(1).Dimension), ...
Name="obs1InLyr")
fullyConnectedLayer(32,Name="obs1PthOutLyr")
];
obs2Path = [
sequenceInputLayer( ...
prod(obsInfo(2).Dimension), ...
Name="obs2InLyr")
fullyConnectedLayer(32,Name="obs2PthOutLyr")
];
obs3Path = [
sequenceInputLayer( ...
prod(obsInfo(3).Dimension), ...
Name="obs3InLyr")
fullyConnectedLayer(32,Name="obs3PthOutLyr")
];
% Concatenate inputs along the first available dimension
comPath = [
concatenationLayer(1,3,Name="comPthInLyr")
reluLayer
lstmLayer(8,OutputMode="sequence",Name="lstm")
fullyConnectedLayer(16)
reluLayer(Name="comPthOutLyr")
];
% Path layers for mean value
% Using tanhLayer & scalingLayer to scale range from (-1,1) to (-10,10)
meanPath = [
fullyConnectedLayer(prod(actInfo(1).Dimension), ...
Name="meanPthInLyr")
tanhLayer
scalingLayer(Name="meanOutLyr", ...
Scale=actInfo(1).UpperLimit)
];
% Path layers for standard deviations
% Using softplus layer to make them nonnegative
stdPath = [
fullyConnectedLayer(prod(actInfo(1).Dimension), ...
Name="stdPthInLyr")
softplusLayer(Name="stdOutLyr")
];
% Assemble dlnetwork object.
net = dlnetwork;
net = addLayers(net,obs1Path);
net = addLayers(net,obs2Path);
net = addLayers(net,obs3Path);
net = addLayers(net,comPath);
net = addLayers(net,meanPath);
net = addLayers(net,stdPath);
% Connect layers.
net = connectLayers(net,"obs1PthOutLyr","comPthInLyr/in1");
net = connectLayers(net,"obs2PthOutLyr","comPthInLyr/in2");
net = connectLayers(net,"obs3PthOutLyr","comPthInLyr/in3");
net = connectLayers(net,"comPthOutLyr","meanPthInLyr/in");
net = connectLayers(net,"comPthOutLyr","stdPthInLyr/in");
% Plot network.
plot(net)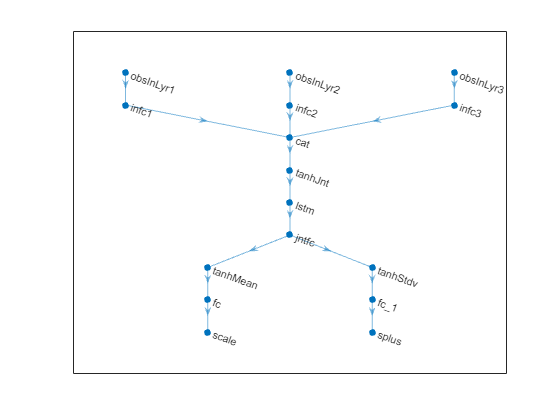
% Initialize network. net = initialize(net); % Display the number of learnable parameters. summary(net)
Initialized: true
Number of learnables: 3.9k
Inputs:
1 'obs1InLyr' Sequence input with 3 dimensions
2 'obs2InLyr' Sequence input with 2 dimensions
3 'obs3InLyr' Sequence input with 1 dimensions
Create the actor with rlContinuousGaussianActor, using the network,
the observations and action specification objects, as well as the names of the network
input layer and the options object.
actor = rlContinuousGaussianActor(net, obsInfo, actInfo, ... ActionMeanOutputNames="meanOutLyr",... ActionStandardDeviationOutputNames="stdOutLyr",... ObservationInputNames=["obs1InLyr","obs2InLyr","obs3InLyr"]);
To return mean value and standard deviations of the Gaussian distribution as a
function of the current observation, use evaluate.
[prob,state] = evaluate(actor, {rand([obsInfo(1).Dimension 1 1]) , ...
rand([obsInfo(2).Dimension 1 1]) , ...
rand([obsInfo(3).Dimension 1 1]) });The result is a cell array with two elements, the first one containing a vector of mean values, and the second containing a vector of standard deviations.
prob{1}ans = 4×1 single column vector
0.0408
0.1472
-0.0644
-0.0433
prob{2}ans = 4×1 single column vector
0.6966
0.6921
0.6795
0.6859
To return an action sampled from the distribution, use
getAction.
act = getAction(actor, {rand(obsInfo(1).Dimension) , ...
rand(obsInfo(2).Dimension) , ...
rand(obsInfo(3).Dimension) });
act{1}ans = 4×1 single column vector
-0.0170
1.8817
0.4884
-0.4666
Calculate the gradients of the sum of the outputs (all the mean values plus all the standard deviations) with respect to the inputs, given a random observation.
gro = gradient(actor,"output-input", ... {rand(obsInfo(1).Dimension) , ... rand(obsInfo(2).Dimension) , ... rand(obsInfo(3).Dimension)} )
gro=3×1 cell array
{3×1 single }
{[-0.0424 0.0307]}
{[ 0.0499]}
The result is a cell array with as many elements as the number of input channels. Each element contains the derivatives of the sum of the outputs with respect to each component of the input channel. Display the gradient with respect to the element of the second channel.
gro{2}ans = 1×2 single row vector
-0.0424 0.0307
Obtain the gradient with respect of five independent sequences, each one made of nine sequential observations.
gro_batch = gradient(actor,"output-input", ... {rand([obsInfo(1).Dimension 5 9]) , ... rand([obsInfo(2).Dimension 5 9]) , ... rand([obsInfo(3).Dimension 5 9])} )
gro_batch=3×1 cell array
{3×1×5×9 single}
{1×2×5×9 single}
{1×1×5×9 single}
Display the derivative of the sum of the outputs with respect to the third observation element of the first input channel, after the seventh sequential observation in the fourth independent batch.
gro_batch{1}(3,1,4,7)ans = single
-0.2679
Set the option to accelerate the gradient computations.
actor = accelerate(actor,true);
Calculate the gradients of the sum of the outputs with respect to the parameters, given a random observation.
grp = gradient(actor,"output-parameters", ... {rand(obsInfo(1).Dimension) , ... rand(obsInfo(2).Dimension) , ... rand(obsInfo(3).Dimension)} )
grp=15×1 cell array
{32×3 single}
{32×1 single}
{32×2 single}
{32×1 single}
{32×1 single}
{32×1 single}
{32×96 single}
{32×8 single}
{32×1 single}
{16×8 single}
{16×1 single}
{ 4×16 single}
{ 4×1 single}
{ 4×16 single}
{ 4×1 single}
Each array within a cell contains the gradient of the sum of the outputs with respect to a group of parameters.
grp_batch = gradient(actor,"output-parameters", ... {rand([obsInfo(1).Dimension 5 9]) , ... rand([obsInfo(2).Dimension 5 9]) , ... rand([obsInfo(3).Dimension 5 9])} )
grp_batch=15×1 cell array
{32×3 single}
{32×1 single}
{32×2 single}
{32×1 single}
{32×1 single}
{32×1 single}
{32×96 single}
{32×8 single}
{32×1 single}
{16×8 single}
{16×1 single}
{ 4×16 single}
{ 4×1 single}
{ 4×16 single}
{ 4×1 single}
If you use a batch of inputs, gradient uses the whole input
sequence (in this case nine steps), and all the gradients with respect to the
independent batch dimensions (in this case five) are added together. Therefore, the
returned gradient has always the same size as the output from getLearnableParameters.
Create observation and action specification objects (or
alternatively use getObservationInfo and
getActionInfo to extract the specification objects from an
environment). For this example, define an observation space made of two channels. The
first channel carries an observation from a continuous four-dimensional space. The
second carries a discrete scalar observation that can be either zero or one. Finally,
the action space consists of a scalar that can be -1,
0, or 1.
obsInfo = [
rlNumericSpec([4 1])
rlFiniteSetSpec([0 1])
];
actInfo = rlFiniteSetSpec([-1 0 1]);To approximate the vector Q-value function within the critic, use a recurrent deep neural network. The output layer must have three elements, each one expressing the value of executing the corresponding action, given the observation.
Create the neural network, defining each network path as an array of layer objects.
Get the dimensions of the observation and action spaces from the environment
specification objects, use sequenceInputLayer as the input layer, and
include an lstmLayer as one of the other network layers.
% First path inPath1 = [ sequenceInputLayer( ... prod(obsInfo(1).Dimension), ... Name="obsInLyr1") fullyConnectedLayer( ... prod(actInfo.Dimension), ... Name="infc1") ]; % Second path inPath2 = [ sequenceInputLayer( ... prod(obsInfo(2).Dimension), ... Name="obsInLyr2") fullyConnectedLayer( ... prod(actInfo.Dimension), ... Name="infc2") ]; % Concatenate inputs along first available dimension. jointPath = [ concatenationLayer(1,2,Name="cct") tanhLayer(Name="tanhJnt") lstmLayer(8,OutputMode="sequence") fullyConnectedLayer(prod(numel(actInfo.Elements))) ];
Assemble dlnetwork object and add layers.
net = dlnetwork; net = addLayers(net,inPath1); net = addLayers(net,inPath2); net = addLayers(net,jointPath);
Connect layers.
net = connectLayers(net,"infc1","cct/in1"); net = connectLayers(net,"infc2","cct/in2");
Plot network.
plot(net)
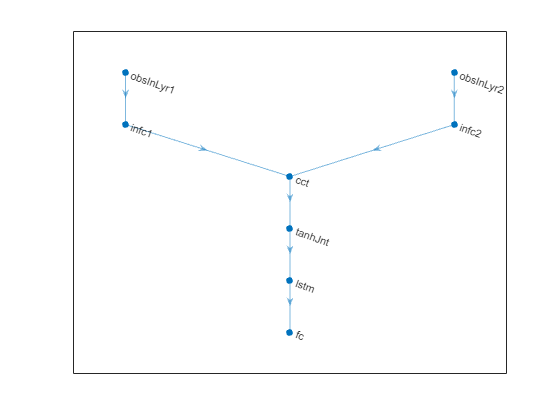
Initialize the network.
net = initialize(net);
Display the number of weights.
summary(net)
Initialized: true
Number of learnables: 386
Inputs:
1 'obsInLyr1' Sequence input with 4 dimensions
2 'obsInLyr2' Sequence input with 1 dimensions
Create the critic with rlVectorQValueFunction, using the network
and the observation and action specification objects.
critic = rlVectorQValueFunction(net,obsInfo,actInfo);
To return the value of the actions as a function of the current observation, use
getValue or evaluate.
val = evaluate(critic, ... {rand(obsInfo(1).Dimension), ... rand(obsInfo(2).Dimension)})
val = 1×1 cell array
{3×1 single}
When you use evaluate, the result is a single-element cell array,
containing a vector with the values of all the possible actions, given the
observation.
val{1}ans = 3×1 single column vector
0.1293
-0.0549
0.0425
Calculate the gradients of the sum of the outputs with respect to the inputs, given a random observation.
gro = gradient(critic,"output-input", ... {rand(obsInfo(1).Dimension) , ... rand(obsInfo(2).Dimension) } )
gro=2×1 cell array
{4×1 single}
{[ 0.0611]}
The result is a cell array with as many elements as the number of input channels. Each element contains the derivative of the sum of the outputs with respect to each component of the input channel. Display the gradient with respect to the element of the second channel.
gro{2}ans = single
0.0611
Obtain the gradient with respect of five independent sequences each one made of nine sequential observations.
gro_batch = gradient(critic,"output-input", ... {rand([obsInfo(1).Dimension 5 9]) , ... rand([obsInfo(2).Dimension 5 9]) } )
gro_batch=2×1 cell array
{4×1×5×9 single}
{1×1×5×9 single}
Display the derivative of the sum of the outputs with respect to the third observation element of the first input channel, after the seventh sequential observation in the fourth independent batch.
gro_batch{1}(3,1,4,7)ans = single
-0.1192
Set the option to accelerate the gradient computations.
critic = accelerate(critic,true);
Calculate the gradients of the sum of the outputs with respect to the parameters, given a random observation.
grp = gradient(critic,"output-parameters", ... {rand(obsInfo(1).Dimension) , ... rand(obsInfo(2).Dimension) } )
grp=9×1 cell array
{[-0.0178 -0.0052 -0.0588 -0.0114]}
{[ -0.0597]}
{[ 0.0873]}
{[ 0.0962]}
{32×2 single }
{32×8 single }
{32×1 single }
{ 3×8 single }
{ 3×1 single }
Each array within a cell contains the gradient of the sum of the outputs with respect to a group of parameters.
grp_batch = gradient(critic,"output-parameters", ... {rand([obsInfo(1).Dimension 5 9]) , ... rand([obsInfo(2).Dimension 5 9]) })
grp_batch=9×1 cell array
{[-2.4487 -2.3282 -2.4941 -2.5877]}
{[ -5.1411]}
{[ 4.4295]}
{[ 9.2137]}
{32×2 single }
{32×8 single }
{32×1 single }
{ 3×8 single }
{ 3×1 single }
If you use a batch of inputs, gradient uses the whole input
sequence (in this case nine steps), and all the gradients with respect to the
independent batch dimensions (in this case five) are added together. Therefore, the
returned gradient always has the same size as the output from getLearnableParameters.
Input Arguments
Function approximator object, specified as one of the following:
rlValueFunctionobject — Value function criticrlQValueFunctionobject — Q-value function criticrlVectorQValueFunctionobject — Multi-output Q-value function critic with a discrete action spacerlContinuousDeterministicActorobject — Deterministic policy actor with a continuous action spacerlDiscreteCategoricalActor— Stochastic policy actor with a discrete action spacerlContinuousGaussianActorobject — Stochastic policy actor with a continuous action spacerlContinuousDeterministicTransitionFunctionobject — Continuous deterministic transition function for a model based agentrlContinuousGaussianTransitionFunctionobject — Continuous Gaussian transition function for a model based agentrlContinuousDeterministicRewardFunctionobject — Continuous deterministic reward function for a model based agentrlContinuousGaussianRewardFunctionobject — Continuous Gaussian reward function for a model based agent.rlIsDoneFunctionobject — Is-done function for a model based agent.
Input data for the function approximator, specified as a cell array with as many
elements as the number of input channels of fcnAppx. In the
following section, the number of observation channels is indicated by
NO.
If
fcnAppxis anrlQValueFunction, anrlContinuousDeterministicTransitionFunctionor anrlContinuousGaussianTransitionFunctionobject, then each of the first NO elements ofinDatamust be a matrix representing the current observation from the corresponding observation channel. They must be followed by a final matrix representing the action.If
fcnAppxis a function approximator object representing an actor or critic (but not anrlQValueFunctionobject),inDatamust contain NO elements, each one being a matrix representing the current observation from the corresponding observation channel.If
fcnAppxis anrlContinuousDeterministicRewardFunction, anrlContinuousGaussianRewardFunction, or anrlIsDoneFunctionobject, then each of the first NO elements ofinDatamust be a matrix representing the current observation from the corresponding observation channel. They must be followed by a matrix representing the action, and finally by NO elements, each one being a matrix representing the next observation from the corresponding observation channel.
Each element of inData must be a matrix of dimension
MC-by-LB-by-LS,
where:
MC corresponds to the dimensions of the associated input channel.
LB is the batch size. To specify a single observation, set LB = 1. To specify a batch of (independent) observations, specify LB > 1. If
inDatahas multiple elements, then LB must be the same for all elements ofinData.LS specifies the sequence length (along the time dimension) for recurrent neural network. If
fcnAppxdoes not use a recurrent neural network, (which is the case of environment function approximators, as they do not support recurrent neural networks) then LS = 1. IfinDatahas multiple elements, then LS must be the same for all elements ofinData.
For more information on input and output formats for recurrent neural networks, see
the Algorithms section of lstmLayer.
Example: {rand(8,3,64,1),rand(4,1,64,1),rand(2,1,64,1)}
Loss function, specified as a function handle to a user-defined function. The user
defined function can either be an anonymous function or a function on the MATLAB path.
The function first input parameter must be a cell array like the one returned from the
evaluation of fcnAppx. For more information, see the description of
outData in evaluate. The
second, optional, input argument of lossFcn contains additional
data that might be needed for the gradient calculation, as described below in
fcnData. For an example of the signature that this function must
have, see Train Reinforcement Learning Policy Using Custom Training Loop.
Additional data for the loss function, specified as any MATLAB data type, typically a structure or cell array. For an example see Train Reinforcement Learning Policy Using Custom Training Loop.
Output Arguments
Value of the gradient, returned as a cell array.
When the type of gradient is from the sum of the outputs with respect to the inputs
of fcnAppx, then grad is a cell array in which
each element contains the gradient of the sum of all the outputs with respect to the
corresponding input channel.
The numerical array in each cell has dimensions D-by-LB-by-LS, where:
D corresponds to the dimensions of the input channel of
fcnAppx.LB is the batch size (length of a batch of independent inputs).
LS is the sequence length (length of the sequence of inputs along the time dimension) for a recurrent neural network. If
fcnAppxdoes not use a recurrent neural network (which is the case of environment function approximators, as they do not support recurrent neural networks), then LS = 1.
When the type of gradient is from the output with respect to the parameters of
fcnAppx, then grad is a cell array in which
each element contains the gradient of the sum of outputs belonging to an output channel
with respect to the corresponding group of parameters. The gradient is calculated using
the whole history of LS inputs, and all the
LB gradients with respect to the
independent input sequences are added together in grad. Therefore,
grad has always the same size as the result from getLearnableParameters.
For more information on input and output formats for recurrent neural networks, see
the Algorithms section of lstmLayer.
Version History
Introduced in R2022agradient is no longer recommended.
Instead of using gradient on a function approximator object, write an appropriate loss function
that takes as arguments both the approximation object and its input. In the loss function
you typically use evaluate to calculate the output and dlgradient to
calculate the gradient. Then call dlfeval,
supplying both the approximator object and it inputs as arguments.
This workflow is shown in the following table.
gradient: Not Recommended | dlfeval and dlgradient: Recommended |
|---|---|
g = gradient(actor,"output-input",u);
g{1}
| g = dlfeval(@myOIGFcn,actor,dlarray(u));
g{1}function g = myOIGFcn(actor,u)
y = evaluate(actor,u);
loss = sum(y{1});
g = dlgradient(loss,u); |
g = gradient(actor,"output-parameters",u);
g{1}
| g = dlfeval(@myOPGFcn,actor,dlarray(u));
g{1}function g = myOIGFcn(actor,u)
y = evaluate(actor,u);
loss = sum(y{1});
g = dlgradient(loss,actor.Learnables);
|
g = gradient(actor,@customLoss23b,u);
g{1}
function loss = customLoss23b(y,varargin)
loss = sum(y{1}.^2); | g = dlfeval(@customLoss24a,actor,dlarray(u));
g{1}function g = customLoss24a(actor,u)
y = evaluate(actor,u);
loss = sum(y{1}.^2);
g = dlgradient(loss,actor.Learnables);
|
For more information, see also accelerate is not recommended.
For more information on using dlarray objects for custom deep learning
training loops, see dlfeval, AcceleratedFunction, dlaccelerate.
For an example, see Train Reinforcement Learning Policy Using Custom Training Loop and Custom Training Loop with Simulink Action Noise.
See Also
Functions
Objects
AcceleratedFunction|rlValueFunction|rlQValueFunction|rlVectorQValueFunction|rlContinuousDeterministicActor|rlDiscreteCategoricalActor|rlContinuousGaussianActor|rlContinuousDeterministicTransitionFunction|rlContinuousGaussianTransitionFunction|rlContinuousDeterministicRewardFunction|rlContinuousGaussianRewardFunction|rlIsDoneFunction
MATLAB Command
You clicked a link that corresponds to this MATLAB command:
Run the command by entering it in the MATLAB Command Window. Web browsers do not support MATLAB commands.
Select a Web Site
Choose a web site to get translated content where available and see local events and offers. Based on your location, we recommend that you select: .
You can also select a web site from the following list
How to Get Best Site Performance
Select the China site (in Chinese or English) for best site performance. Other MathWorks country sites are not optimized for visits from your location.
Americas
- América Latina (Español)
- Canada (English)
- United States (English)
Europe
- Belgium (English)
- Denmark (English)
- Deutschland (Deutsch)
- España (Español)
- Finland (English)
- France (Français)
- Ireland (English)
- Italia (Italiano)
- Luxembourg (English)
- Netherlands (English)
- Norway (English)
- Österreich (Deutsch)
- Portugal (English)
- Sweden (English)
- Switzerland
- United Kingdom (English)