fitsemigraph
Label data using semi-supervised graph-based method
Syntax
Description
fitsemigraph creates a semi-supervised graph-based model
given labeled data, labels, and unlabeled data. The returned model contains the fitted labels
for the unlabeled data and the corresponding scores. This model can also predict labels for
unseen data using the predict object function. For more information on
the different labeling algorithms, see Algorithms.
Mdl = fitsemigraph(Tbl,ResponseVarName,UnlabeledTbl)Tbl, where
Tbl.ResponseVarName contains the labels for the labeled data, and
returns fitted labels for the unlabeled data in UnlabeledTbl. The
function stores the fitted labels and the corresponding scores in the
FittedLabels and LabelScores properties of the
object Mdl, respectively.
Mdl = fitsemigraph(Tbl,formula,UnlabeledTbl)formula to specify the response variable (vector of labels) and
the predictor variables to use among the variables in Tbl. The
function uses these variables to label the data in
UnlabeledTbl.
Mdl = fitsemigraph(Tbl,Y,UnlabeledTbl)Tbl and the labels in
Y to label the data in UnlabeledTbl.
Mdl = fitsemigraph(X,Y,UnlabeledX)X and the labels in Y
to label the data in UnlabeledX.
Mdl = fitsemigraph(___,Name,Value)
Examples
Fit labels to unlabeled data by using a semi-supervised graph-based method.
Randomly generate 60 observations of labeled data, with 20 observations in each of three classes.
rng('default') % For reproducibility labeledX = [randn(20,2)*0.25 + ones(20,2); randn(20,2)*0.25 - ones(20,2); randn(20,2)*0.5]; Y = [ones(20,1); ones(20,1)*2; ones(20,1)*3];
Visualize the labeled data by using a scatter plot. Observations in the same class have the same color. Notice that the data is split into three clusters with very little overlap.
scatter(labeledX(:,1),labeledX(:,2),[],Y,'filled') title('Labeled Data')

Randomly generate 300 additional observations of unlabeled data, with 100 observations per class. For the purposes of validation, keep track of the true labels for the unlabeled data.
unlabeledX = [randn(100,2)*0.25 + ones(100,2);
randn(100,2)*0.25 - ones(100,2);
randn(100,2)*0.5];
trueLabels = [ones(100,1); ones(100,1)*2; ones(100,1)*3];Fit labels to the unlabeled data by using a semi-supervised graph-based method. The function fitsemigraph returns a SemiSupervisedGraphModel object whose FittedLabels property contains the fitted labels for the unlabeled data and whose LabelScores property contains the associated label scores.
Mdl = fitsemigraph(labeledX,Y,unlabeledX)
Mdl =
SemiSupervisedGraphModel with properties:
FittedLabels: [300×1 double]
LabelScores: [300×3 double]
ClassNames: [1 2 3]
ResponseName: 'Y'
CategoricalPredictors: []
Method: 'labelpropagation'
Properties, Methods
Visualize the fitted label results by using a scatter plot. Use the fitted labels to set the color of the observations, and use the maximum label scores to set the transparency of the observations. Observations with less transparency are labeled with greater confidence. Notice that observations that lie closer to the cluster boundaries are labeled with more uncertainty.
maxLabelScores = max(Mdl.LabelScores,[],2); rescaledScores = rescale(maxLabelScores,0.05,0.95); scatter(unlabeledX(:,1),unlabeledX(:,2),[],Mdl.FittedLabels,'filled', ... 'MarkerFaceAlpha','flat','AlphaData',rescaledScores); title('Fitted Labels for Unlabeled Data')

Determine the accuracy of the labeling by using the true labels for the unlabeled data.
numWrongLabels = sum(trueLabels ~= Mdl.FittedLabels)
numWrongLabels = 10
Only 10 of the 300 observations in unlabeledX are mislabeled.
Fit labels to unlabeled data by using a semi-supervised graph-based method. Specify the type of nearest neighbor graph.
Load the patients data set. Create a table from the variables Distolic, Gender, and so on. For each observation, or row in the table, treat the Smoker value as the label for that observation.
load patients
Tbl = table(Diastolic,Gender,Height,Systolic,Weight,Smoker);Suppose only 20% of the observations are labeled. To recreate this scenario, randomly sample 20 labeled observations and store them in the table unlabeledTbl. Remove the label from the rest of the observations and store them in the table unlabeledTbl. To verify the accuracy of the label fitting at the end of the example, retain the true labels for the unlabeled data in the variable trueLabels.
rng('default') % For reproducibility of the sampling [labeledTbl,Idx] = datasample(Tbl,20,'Replace',false); unlabeledTbl = Tbl; unlabeledTbl(Idx,:) = []; trueLabels = unlabeledTbl.Smoker; unlabeledTbl.Smoker = [];
Fit labels to the unlabeled data by using a semi-supervised graph-based method. Use a mutual type of nearest neighbor graph, where two points are connected when they are nearest neighbors of each other. Specify to standardize the numeric predictors. The function fitsemigraph returns an object whose FittedLabels property contains the fitted labels for the unlabeled data.
Mdl = fitsemigraph(labeledTbl,'Smoker',unlabeledTbl,'KNNGraphType','mutual', ... 'Standardize',true); fittedLabels = Mdl.FittedLabels;
Identify the observations that are incorrectly labeled by comparing the stored true labels for the unlabeled data to the fitted labels returned by the semi-supervised graph-based method.
wrongIdx = (trueLabels ~= fittedLabels); wrongTbl = unlabeledTbl(wrongIdx,:);
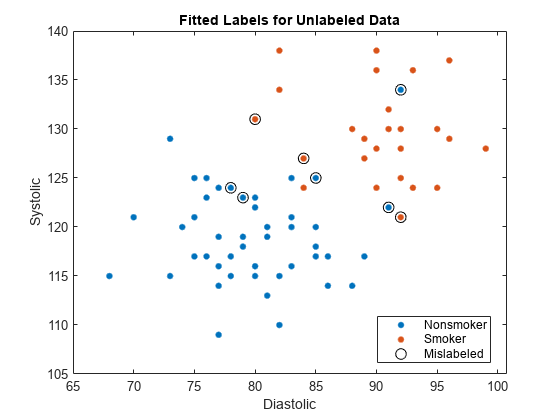
Visualize the fitted label results for the unlabeled data. Mislabeled observations are circled in the plot.
gscatter(unlabeledTbl.Diastolic,unlabeledTbl.Systolic, ... fittedLabels) hold on plot(wrongTbl.Diastolic,wrongTbl.Systolic, ... 'ko','MarkerSize',8) xlabel('Diastolic') ylabel('Systolic') legend('Nonsmoker','Smoker','Mislabeled') title('Fitted Labels for Unlabeled Data')
Input Arguments
Labeled sample data, specified as a table. Each row of Tbl
corresponds to one observation, and each column corresponds to one predictor.
Optionally, Tbl can contain one additional column for the response
variable (vector of labels). Multicolumn variables, cell arrays other than cell arrays
of character vectors, and ordinal categorical variables are not supported.
If Tbl contains the response variable, and you want to use all
remaining variables in Tbl as predictors, then specify the response
variable using ResponseVarName.
If Tbl contains the response variable, and you want to use only
a subset of the remaining variables in Tbl as predictors, specify a
formula using formula.
If Tbl does not contain the response variable, specify a
response variable using Y. The length of the response variable and
the number of rows in Tbl must be equal.
Data Types: table
Unlabeled sample data, specified as a table. Each row of
UnlabeledTbl corresponds to one observation, and each column
corresponds to one predictor. UnlabeledTbl must contain the same
predictors as those contained in Tbl.
Data Types: table
Response variable name, specified as the name of a variable in
Tbl. The response variable contains the class labels for the
sample data in Tbl.
You must specify ResponseVarName as a character vector or string
scalar. For example, if the response variable Y is stored as
Tbl.Y, then specify it as 'Y'. Otherwise, the
software treats all columns of Tbl, including Y,
as predictors.
The response variable must be a categorical, character, or string array, a logical
or numeric vector, or a cell array of character vectors. If Y is a
character array, then each element of the response variable must correspond to one row
of the array.
A good practice is to specify the order of the classes by using the
ClassNames name-value pair argument.
Data Types: char | string
Explanatory model of the response variable and a subset of the predictor variables,
specified as a character vector or string scalar in the form
'Y~X1+X2+X3'. In this form, Y represents the
response variable, and X1, X2, and
X3 represent the predictor variables.
To specify a subset of variables in Tbl as predictors, use a
formula. If you specify a formula, then the software does not use any variables in
Tbl that do not appear in formula.
The variable names in the formula must be both variable names in Tbl
(Tbl.Properties.VariableNames) and valid MATLAB® identifiers. You can verify the variable names in Tbl by
using the isvarname function. If the variable names
are not valid, then you can convert them by using the matlab.lang.makeValidName function.
Data Types: char | string
Class labels, specified as a numeric, categorical, or logical vector, a character or string array, or a cell array of character vectors.
If
Yis a character array, then each element of the class labels must correspond to one row of the array.The length of
Ymust be equal to the number of rows inTblorX.A good practice is to specify the class order by using the
ClassNamesname-value pair argument.
Data Types: single | double | categorical | logical | char | string | cell
Labeled predictor data, specified as a numeric matrix.
Each row of X corresponds to one observation, and each column
corresponds to one predictor.
The length of Y and the number of rows in
X must be equal.
To specify the names of the predictors in the order of their appearance in
X, use the PredictorNames name-value pair
argument.
Data Types: single | double
Unlabeled predictor data, specified as a numeric matrix. Each row of
UnlabeledX corresponds to one observation, and each column
corresponds to one predictor. UnlabeledX must have the same
predictors as X, in the same order.
Data Types: single | double
Note
The software treats NaN, empty character vector
(''), empty string (""),
<missing>, and <undefined> elements as
missing data. The software removes rows of the predictor data (observations) with missing
values.
Name-Value Arguments
Specify optional pairs of arguments as
Name1=Value1,...,NameN=ValueN, where Name is
the argument name and Value is the corresponding value.
Name-value arguments must appear after other arguments, but the order of the
pairs does not matter.
Before R2021a, use commas to separate each name and value, and enclose
Name in quotes.
Example: fitsemigraph(Tbl,'Y',UnlabeledTbl,'Method','labelspreading','IterationLimit',2e3)
specifies to use a label spreading labeling technique and run a maximum of 2000
iterations.
Labeling Algorithm Options
Labeling technique, specified as the comma-separated pair consisting of
'Method' and one of these values.
| Value | Description | Method-Specific Name-Value Pair Arguments |
|---|---|---|
'labelpropagation' | Iteratively propagate labels across the nodes in the similarity graph. For more information, see Label Propagation. |
|
'labelpropagationexact' | Use an exact formula to propagate labels. For more information, see Label Propagation. | None |
'labelspreading' | Iteratively spread labels across the nodes in the similarity graph. For more information, see Label Spreading. |
|
'labelspreadingexact' | Use an exact formula to spread labels. For more information, see Label Spreading. |
|
Example: 'Method','labelspreading'
Data Types: char | string
Relative weight of neighboring labels to the initial label for labeled
observations in X or Tbl, specified as the
comma-separated pair consisting of 'Alpha' and a scalar value in
the range (0,1). A value close to 0 indicates that fitsemigraph
treats labels of initially labeled observations almost like ground truth. A value
close to 1 indicates that fitsemigraph treats labels of initially
labeled observations almost like noise.
Note
This argument is valid only when the Method value is
'labelspreading' or
'labelspreadingexact'.
Example: 'Alpha',0.05
Data Types: single | double
Maximum number of iterations, specified as the comma-separated pair consisting of
'IterationLimit' and a positive integer scalar. The
fitsemigraph function returns Mdl, which
contains the fitted labels and scores, when this limit is reached, even if the
algorithm does not converge.
Note
This argument is valid only when the Method value is
'labelpropagation' or
'labelspreading'.
Example: 'IterationLimit',2e3
Data Types: single | double
Tolerance for score absolute difference in subsequent iterations, specified as the
comma-separated pair consisting of 'Tolerance' and a nonnegative
scalar.
Note
This argument is valid only when the Method value is
'labelpropagation' or
'labelspreading'.
Example: 'Tolerance',1e-4
Data Types: single | double
Classification Options
Categorical predictors list, specified as one of the values in this table.
| Value | Description |
|---|---|
| Vector of positive integers |
Each entry in the vector is an index value indicating that the corresponding predictor is
categorical. The index values are between 1 and If |
| Logical vector |
A |
| Character matrix | Each row of the matrix is the name of a predictor variable. The names must match the entries in PredictorNames. Pad the names with extra blanks so each row of the character matrix has the same length. |
| String array or cell array of character vectors | Each element in the array is the name of a predictor variable. The names must match the entries in PredictorNames. |
"all" | All predictors are categorical. |
By default, if the predictor data is in a table, fitsemigraph
assumes that a variable is categorical if it is a logical vector, categorical vector,
character array, string array, or cell array of character vectors. Ordinal categorical
variables are not supported. If the predictor data is a matrix,
fitsemigraph assumes that all predictors are continuous. To
identify any other predictors as categorical predictors, specify them by using the
'CategoricalPredictors' name-value pair argument.
fitsemigraph encodes categorical variables as numeric
variables by assigning a positive integer value to each category. When you use
categorical predictors, ensure that you use an appropriate distance metric
(Distance).
Example: 'CategoricalPredictors','all'
Data Types: single | double | logical | char | string | cell
Names of the classes to use for labeling, specified as the comma-separated pair
consisting of 'ClassNames' and a categorical, character, or string
array, a logical or numeric vector, or a cell array of character vectors.
ClassNames must have the same data type as
Y.
If ClassNames is a character array, then each element must
correspond to one row of the array.
Use 'ClassNames' to:
Order the classes.
Specify the order of any input or output argument dimension that corresponds to the class order. For example, use
'ClassNames'to specify the column order of classification scores inMdl.LabelScores.Select a subset of classes for labeling. For example, suppose that the set of all distinct class names in
Yis{'a','b','c'}. To use observations from classes'a'and'c'only, specify'ClassNames',{'a','c'}.
The default value for ClassNames is the set of all distinct
class names in Y.
Example: 'ClassNames',{'b','g'}
Data Types: categorical | char | string | logical | single | double | cell
Predictor variable names, specified as the comma-separated pair consisting of
'PredictorNames' and a string array of unique names or cell array
of unique character vectors. The functionality of 'PredictorNames'
depends on the way you supply the predictor data.
If you supply
X,Y, andUnlabeledX, then you can use'PredictorNames'to assign names to the predictor variables inXandUnlabeledX.The order of the names in
PredictorNamesmust correspond to the column order ofX. That is,PredictorNames{1}is the name ofX(:,1),PredictorNames{2}is the name ofX(:,2), and so on. Also,size(X,2)andnumel(PredictorNames)must be equal.By default,
PredictorNamesis{'x1','x2',...}.
If you supply
TblandUnlabeledTbl, then you can use'PredictorNames'to choose which predictor variables to use. That is,fitsemigraphuses only the predictor variables inPredictorNamesand the response variable to label the unlabeled data.PredictorNamesmust be a subset ofTbl.Properties.VariableNamesand cannot include the name of the response variable.By default,
PredictorNamescontains the names of all predictor variables.A good practice is to specify the predictors using either
'PredictorNames'orformula, but not both.
Example: 'PredictorNames',{'SepalLength','SepalWidth','PetalLength','PetalWidth'}
Data Types: string | cell
Response variable name, specified as the comma-separated pair consisting of
'ResponseName' and a character vector or string scalar.
If you supply
Y, then you can use'ResponseName'to specify a name for the response variable.If you supply
ResponseVarNameorformula, then you cannot use'ResponseName'.
Example: 'ResponseName','response'
Data Types: char | string
Flag to standardize the predictor data, specified as the comma-separated pair
consisting of 'Standardize' and a numeric or logical
0 (false) or 1
(true). If you set 'Standardize',true, the
software combines the labeled and unlabeled predictor data, and then centers and
scales each numeric predictor variable by the corresponding column mean and standard
deviation. The software does not standardize the categorical predictors.
Example: 'Standardize',true
Data Types: double | logical
Distance Metric Options
Distance metric, specified as the comma-separated pair consisting of
'Distance' and a character vector or string scalar.
If all the predictor variables are continuous (numeric) variables, then you can specify one of these distance metrics.
Value Description 'euclidean'Euclidean distance
'seuclidean'Standardized Euclidean distance — Each coordinate difference between observations is scaled by dividing by the corresponding element of the standard deviation
S = std(PD,'omitnan'), wherePDis the predictor data, both labeled and unlabeled. To specify another scale parameter, use the'Scale'name-value pair argument.'mahalanobis'Mahalanobis distance — By default, the distance is computed using
C = cov(PD,'omitrows'), the covariance ofPD. To change the value of the covariance matrix, use the'Cov'name-value pair argument.'minkowski'Minkowski distance — The default exponent is 2. To specify a different exponent, use the
'P'name-value pair argument.'chebychev'Chebychev distance (maximum coordinate difference)
'cityblock'City block distance
'correlation'One minus the sample correlation between observations (treated as sequences of values)
'cosine'One minus the cosine of the included angle between observations (treated as vectors)
'spearman'One minus the sample Spearman's rank correlation between observations (treated as sequences of values)
Note
If you specify one of these distance metrics and the predictor data includes categorical predictors, then the software treats each categorical predictor as a numeric variable for the distance computation, with each category represented by a positive integer. The
Distancevalue does not affect theCategoricalPredictorsproperty of the trained model.If all the predictor variables are categorical variables, then you can specify one of these distance metrics.
Value Description 'hamming'Hamming distance, which is the percentage of coordinates that differ
'jaccard'One minus the Jaccard coefficient, which is the percentage of nonzero coordinates that differ
Note
If you specify one of these distance metrics and the predictor data includes continuous (numeric) predictors, then the software treats each continuous predictor as a categorical variable for the distance computation. The
Distancevalue does not affect theCategoricalPredictorsproperty of the trained model.If the predictor variables are a mix of continuous (numeric) and categorical variables, then you can specify one of these distance metrics.
Value Description 'goodall3'Modified Goodall distance
'ofd'Occurrence frequency distance
The default value is 'euclidean' if all the predictor variables
are continuous, and 'goodall3' if any of the predictor variables
are categorical. For more information on the various distance metrics, see Distance Metrics.
Example: 'Distance','ofd'
Data Types: char | string
Scale parameter for the standardized Euclidean distance metric, specified as the
comma-separated pair consisting of 'Scale' and a nonnegative
vector. The length of Scale is equal to the number of predictors.
Each coordinate difference between two observations is scaled by the corresponding
element of Scale.
The default scale parameter is std(PD,'omitnan'), where
PD is the predictor data, both labeled and unlabeled.
Note
This argument is valid only if Distance is
'seuclidean'.
Example: 'Scale',iqr(X)
Data Types: single | double
Covariance matrix for the Mahalanobis distance metric, specified as the
comma-separated pair consisting of 'Cov' and a
p-by-p positive definite matrix, where
p is the number of predictors.
The default covariance matrix is cov(PD,'omitrows'), where
PD is the predictor data, both labeled and unlabeled.
Note
This argument is valid only if Distance is
'mahalanobis'.
Example: 'Cov',eye(3)
Data Types: single | double
Exponent for the Minkowski distance metric, specified as the comma-separated pair
consisting of 'P' and a positive scalar.
Note
This argument is valid only if Distance is
'minkowski'.
Example: 'P',3
Data Types: single | double
Graph Options
Type of similarity graph used in the labeling algorithm, specified as the
comma-separated pair consisting of 'SimilarityGraph' and one of
these values.
| Value | Description | Graph-Specific Name-Value Pair Arguments |
|---|---|---|
'knn' | Construct the graph using nearest neighbors. |
|
'epsilon' | Construct the graph by using radius search. You must specify a value
for Radius if you use this option. | 'Radius' — Search radius for the nearest neighbors
used to construct the similarity graph |
For more information, see Similarity Graph.
Example: 'SimilarityGraph','epsilon','Radius',2
Data Types: char | string
Number of nearest neighbors used to construct the similarity graph, specified as
the comma-separated pair consisting of 'NumNeighbors' and a
positive integer scalar.
The default number of neighbors is log(n), where
n is the number of observations in the predictor data, both
labeled and unlabeled.
Note
This argument is valid only if SimilarityGraph is
'knn'.
Example: 'NumNeighbors',10
Data Types: single | double
Type of nearest neighbor graph, specified as the comma-separated pair consisting
of 'KNNGraphType' and one of these values.
| Value | Description |
|---|---|
'complete' | Connects two points i and j, when either i is a nearest neighbor of j or j is a nearest neighbor of i. This option leads to a denser representation of the similarity matrix. |
'mutual' | Connects two points i and j, when i is a nearest neighbor of j and j is a nearest neighbor of i. This option leads to a sparser representation of the similarity matrix. |
Note
This argument is valid only if SimilarityGraph is
'knn'.
Example: 'KNNGraphType','mutual'
Data Types: char | string
Search radius for the nearest neighbors used to construct the similarity graph,
specified as the comma-separated pair consisting of 'Radius' and a
nonnegative scalar.
Note
You must specify this argument if SimilarityGraph is
'epsilon'.
Example: 'Radius',5
Data Types: single | double
Scale factor for the kernel, specified as the comma-separated pair consisting of
'KernelScale' and 'auto' or a positive scalar.
The software uses the scale factor to transform distances to similarity
measures.
The
'auto'option is supported only for the'euclidean'and'seuclidean'distance metrics.If you specify
'auto', then the software selects an appropriate scale factor using a heuristic procedure. This heuristic procedure uses subsampling, so estimates can vary from one call to another. To reproduce results, set a random number seed usingrngbefore callingfitsemigraph.
Example: 'KernelScale','auto'
Data Types: single | double | char | string
Output Arguments
Semi-supervised graph-based classifier, returned as a SemiSupervisedGraphModel object. Use dot notation to access the object
properties. For example, to get the fitted labels for the unlabeled data and their
corresponding scores, enter Mdl.FittedLabels and
Mdl.LabelScores, respectively.
More About
A distance metric is a function that defines a distance between two
observations. fitsemigraph supports various distance metrics for
continuous (numeric) predictors, categorical predictors, and a mix of continuous and
categorical predictors.
Given an mx-by-n data matrix X, which is treated as mx (1-by-n) row vectors x1, x2, ..., xmx, and an my-by-n data matrix Y, which is treated as my (1-by-n) row vectors y1, y2, ...,ymy, the various distances between the vector xs and yt are defined as follows:
Distance metrics for continuous (numeric) variables
Euclidean distance
The Euclidean distance is a special case of the Minkowski distance, where p = 2.
Standardized Euclidean distance
where V is the n-by-n diagonal matrix whose jth diagonal element is (S(j))2, where S is a vector of scaling factors for each dimension.
Mahalanobis distance
where C is the covariance matrix.
Minkowski distance
For the special case of p = 1, the Minkowski distance gives the city block distance. For the special case of p = 2, the Minkowski distance gives the Euclidean distance. For the special case of p = ∞, the Minkowski distance gives the Chebychev distance.
Chebychev distance
The Chebychev distance is a special case of the Minkowski distance, where p = ∞.
City block distance
The city block distance is a special case of the Minkowski distance, where p = 1.
Correlation distance
where
and
Cosine distance
Spearman distance is one minus the sample Spearman's rank correlation between observations (treated as sequences of values):
where
Distance metrics for categorical variables
Hamming distance
The Hamming distance is the percentage of coordinates that differ.
Jaccard distance is one minus the Jaccard coefficient, which is the percentage of nonzero coordinates that differ:
Distance metrics for a mix of continuous (numeric) and categorical variables
Modified Goodall distance
This distance is a variant of the Goodall distance, which assigns a small distance if the matching values are infrequent regardless of the frequencies of the other values. For mismatches, the distance contribution of the predictor is 1/(number of variables).
Occurrence frequency distance
For a match, the occurrence frequency distance assigns zero distance. For a mismatch, the occurrence frequency distance assigns a higher distance on a less frequent value and a lower distance on a more frequent value.
A similarity graph models the local neighborhood relationships between observations in the predictor data, both labeled and unlabeled, as an undirected graph. The nodes in the graph represent observations, and the edges, which are directionless, represent the connections between the observations.
If the pairwise distance Disti,j between any two nodes i and j is positive (or larger than a certain threshold), then the similarity graph connects the two nodes using an edge. The edge between the two nodes is weighted by the pairwise similarity Si,j, where , for a specified kernel scale σ value.
fitsemigraph supports these two methods of constructing a similarity graph:
Nearest neighbor method (if
SimilarityGraphis'knn'(default)):fitsemigraphconnects observations in the predictor data, both labeled and unlabeled, that are nearest neighbors.Use the
'NumNeighbors'name-value pair argument to specify the number of nearest neighbors.Use the
'KNNGraphType'name-value pair argument to specify whether to make a'complete'or'mutual'connection of points.
Radius search method (if
SimilarityGraphis'epsilon'):fitsemigraphconnects observations whose pairwise distances are smaller than the specified search radius. You must specify the search radius for nearest neighbors used to construct the similarity graph by using the'Radius'name-value pair argument.
A similarity matrix is a matrix representation of a similarity graph. The n-by-n matrix contains pairwise similarity values between connected nodes in the similarity graph. The similarity matrix of a graph is also called an adjacency matrix.
The similarity matrix is symmetric because the edges of the similarity graph are
directionless. A value of Si,j =
0 means that nodes i and j of the
similarity graph are not connected.
Algorithms
The software constructs a similarity graph (SimilarityGraph) with
both labeled and unlabeled observations as nodes, and distributes label information from
labeled observations to unlabeled observations by using either label propagation or label
spreading.
To propagate labels across the nodes in the similarity graph, the iterative label
propagation algorithm (where Method is
'labelpropagation') follows these steps:
Initialize an n-by-K matrix F(0), where n is the number of nodes (or observations) and K is the number of classes.
The first l rows correspond to labeled observations. Each row contains a 1 in the column corresponding to the true class label for that observation, and a 0 in every other column.
The last u rows correspond to the unlabeled observations, and contain a 0 in all columns.
At iteration t (starting with t = 1), update the F matrix by using the probabilistic transition matrix P, so that , where .
Pi,j is the probability of transmitting label information from node i to node j.
Si,j is the weight of the edge between node i and node j. For the definition of Si,j, see Similarity Graph.
To complete iteration t, clamp the labels for the labeled observations. That is, keep the first l rows of F(t) equal to their initial values in F(0).
Repeat the second and third steps until the F values converge. You can use the
IterationLimitandTolerancevalues to control the convergence.The final F matrix corresponds to the scores for the labeled data and the unlabeled data (
LabelScores). For each observation, or row in F, the column with the maximum score corresponds to the fitted class label (FittedLabels).
For more details, see [2].
Instead of using an iterative label propagation algorithm, you can use an exact method
for label propagation (where Method is
'labelpropagationexact'). In this case, the
u-by-K matrix of label information for the unlabeled
data is FU = (I –
PUU)-1PULFL where:
I is the identity matrix.
PUU and PUL are the labeled (L) and unlabeled (U) submatrices of P such that .
FL is the l-by-K matrix of label information for the labeled data. Each row contains a 1 in the column corresponding to the true class label for that observation, and a 0 in every other column.
The FU matrix corresponds to the
scores for the unlabeled data (LabelScores). For
each observation, or row in FU, the column with
the maximum score corresponds to the fitted class label (FittedLabels).
For more details, see [3].
To spread labels across the nodes in the similarity graph, the iterative spreading
propagation algorithm (where Method is
'labelspreading') follows these steps:
Create an n-by-K matrix Y, where n is the number of nodes (or observations) and K is the number of classes.
The first l rows correspond to labeled observations. Each row contains a 1 in the column corresponding to the true class label for that observation, and a 0 in every other column.
The last u rows correspond to the unlabeled observations, and contain a 0 in all columns.
Create a matrix A that is a normalized version of the n-by-n similarity matrix S, with pairwise similarity values Si,j as defined in Similarity Graph. Let A = D-1/2SD-1/2, where D is the n-by-n diagonal matrix .
At iteration t (starting with t = 1), update the F matrix by using the matrix A and the neighboring label weight parameter α (
Alpha), so that F(t) = αAF(t – 1) + (1 – α)Y. Let F(0) equal Y.Repeat the third step until the F values converge. You can use the
IterationLimitandTolerancevalues to control the convergence.Take the limit of the sequence . This final matrix corresponds to the scores for the labeled data and the unlabeled data (
LabelScores). For each observation, or row in the matrix, the column with the maximum score corresponds to the fitted class label (FittedLabels).
Instead of using an iterative label spreading algorithm, you can use an exact method for
label spreading (where Method is
'labelspreadingexact'). In this case, the
n-by-K matrix of label information for the labeled
and unlabeled data is F = (I –
αA)-1Y, where I is the identity matrix. The
F matrix corresponds to the scores for the labeled data and the
unlabeled data (LabelScores). For
each observation, or row in F, the column with the maximum score
corresponds to the fitted class label (FittedLabels).
For more details, see [1].
References
[1] Zhou, Dengyong, Olivier Bousquet, Thomas Navin Lal, Jason Weston, and Bernhard Schölkopf. “Learning with Local and Global Consistency.” Advances in Neural Information Processing Systems 16 (NIPS). 2003.
[2] Zhu, Xiaojin, and Zoubin Ghahramani. “Learning from Labeled and Unlabeled Data with Label Propagation.” CMU CALD tech report CMU-CALD-02-107. 2002.
[3] Zhu, Xiaojin, Zoubin Ghahramani, and John Lafferty. “Semi-Supervised Learning Using Gaussian Fields and Harmonic Functions.” The Twentieth International Conference on Machine Learning (ICML). 2003.
Version History
Introduced in R2020b
MATLAB Command
You clicked a link that corresponds to this MATLAB command:
Run the command by entering it in the MATLAB Command Window. Web browsers do not support MATLAB commands.
Select a Web Site
Choose a web site to get translated content where available and see local events and offers. Based on your location, we recommend that you select: .
You can also select a web site from the following list
How to Get Best Site Performance
Select the China site (in Chinese or English) for best site performance. Other MathWorks country sites are not optimized for visits from your location.
Americas
- América Latina (Español)
- Canada (English)
- United States (English)
Europe
- Belgium (English)
- Denmark (English)
- Deutschland (Deutsch)
- España (Español)
- Finland (English)
- France (Français)
- Ireland (English)
- Italia (Italiano)
- Luxembourg (English)
- Netherlands (English)
- Norway (English)
- Österreich (Deutsch)
- Portugal (English)
- Sweden (English)
- Switzerland
- United Kingdom (English)