Deep Learning workflow for computational pathology
Deep Learning workflow for computational pathology
This repository shows an example of how to use MATLAB to manipulate very large images in the context of computational pathology (CPATH).
It is structured in two parts:
- How to handle very large images such as whole slide images (WSIs)
- How to pre- and post-process histology images.
Part 1: Handling gigapixel-sized WSIs
The first part of this example shows how to read, display, explore, and organize WSIs (and their patches) in MATLAB. Thanks to the recently introduced blockedImage object, it is now possible to handle very large images without running out of memory. A blockedImage is an image made from discrete blocks (patches), which can be organized and managed using a blockedImageDatastore object and displayed using bigimageshow.
Part 2: Useful pre- and post-processing operations on WSIs in MATLAB
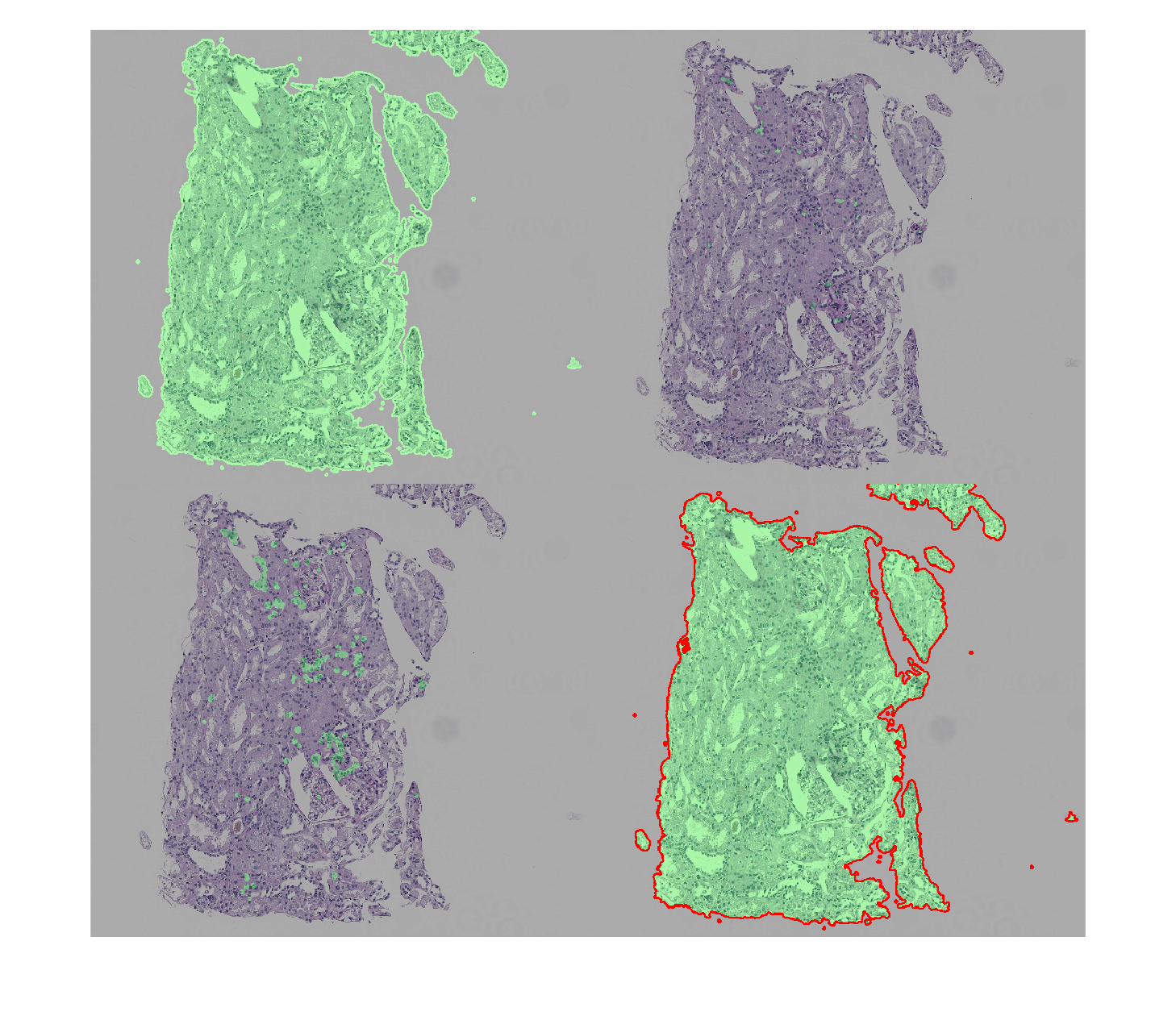
Since the goal of using deep learning techniques in CPATH is to produce solutions that are clinically translatable, i.e., capable of working across large patient populations, it is advisable to deal with some of the most likely WSI artifacts upfront, thereby increasing the abilities of the resulting model to generalize over image artifacts found in other test sets.
The second part of this example shows examples of preprocessing operations to handle commonly found artifacts in histopathology images as well as postprocessing morphological operations for improving the quality of the results at pixel level. Essentially, this example should help the medical image analysis community to create an image analysis pipeline for WSIs (and, as bonus, the ability to reproduce the code and examples described in a recent paper on this topic) using MATLAB.
It highlights the usefulness of MATLAB (and Image Processing Toolbox) functions such as:
- Image thresholding and filtering:
imbinarize,bwareafilt, andimlincomb - Morphological image processing operations:
imclose,imopen,imdilate,imerode,imfill, andstrel - Feature extraction:
bwlabelandregionprops - Visualization:
montage,imoverlay,plotandrectangle
Requirements
- MATLAB 2021a or later
- Image Processing Toolbox
Suggested steps
- Download or clone the repository.
- Open MATLAB.
- Ensure that the files
AT2Scan.jpgandFakePred2.jpgcontaining the test images1 for Part 2 are in the same folder as thecpath_matlab.mlxfile. - Run the
cpath_matlab.mlxscript and inspect results.
Additional remarks
- You are encouraged to expand and adapt the example to your needs.
- The image used for Part 1 is part of MATLAB.
Notes
[1] These images are publicly available at (https://github.com/BHSmith3/Image-Cleaning).
Cite As
Oge Marques (2026). Deep Learning workflow for computational pathology (https://github.com/ogemarques/cpath-matlab/releases/tag/v1.0), GitHub. Retrieved .
MATLAB Release Compatibility
Platform Compatibility
Windows macOS LinuxTags
Discover Live Editor
Create scripts with code, output, and formatted text in a single executable document.
| Version | Published | Release Notes | |
|---|---|---|---|
| 1.0 |